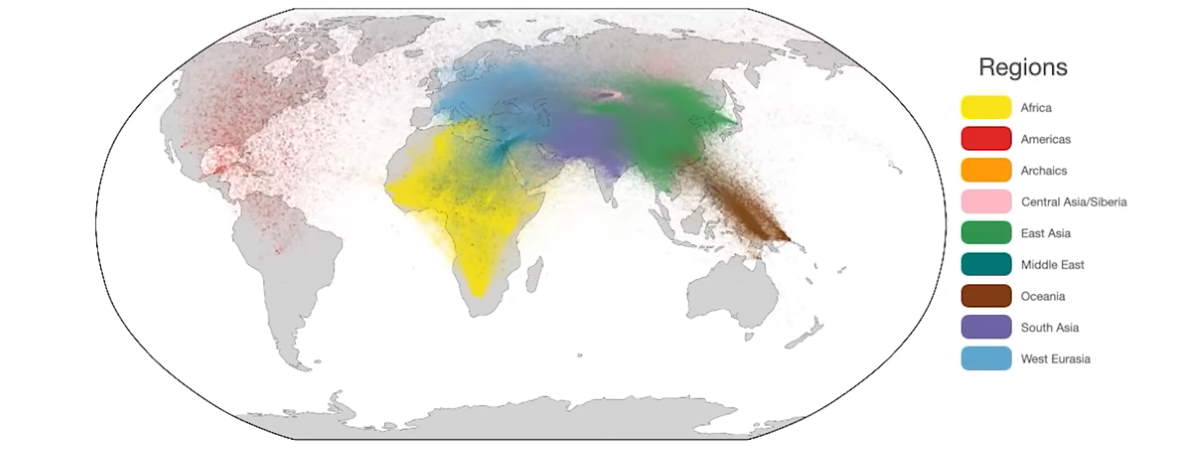
Researchers from the Big Data Institute at the University of Oxford in England used DNA analysis to create the largest human family tree ever, dating back 100,000 years. They published their study titled “A unified genealogy of modern and ancient genomes” in the journal Science.
The characterization of modern and ancient human genome sequences always reveals a little more about the history of our evolutionary past. Since the year 2000, research in human genetics has developed considerably. For example, new techniques for analyzing ancient DNA have made it possible to discover that humans had interbred with Neanderthals. But, the problem was that huge amounts of new information have been generated, so much so that scientists have not been able until now, to process it properly.
Also, although to date thousands of modern and ancient human genomes have been generated, differences in methods and data quality were making it very complicated to compare and/or combine.
These genomic datasets are highly heterogeneous: samples from diverse geographic locations, times, and populations are processed, sequenced, and analyzed using a variety of techniques. The resulting datasets contain true variation, but also complex patterns of misses and errors that make it difficult to get the most complete view of human genomic variation.
Now, researchers from the Big Data Institute (BDI) succeeded in solving this problem: they created algorithms making it possible to combine different databases, and to integrate ancient and modern genomes. It was these algorithms that allowed them to build the structure of what they described as a “human genealogy”, the largest ever human family tree.
The new algorithm applies a tree recording method to ancient and modern human genomes to generate a unified human genealogy. Now, BDI researchers can easily combine data from multiple sources, even use missing and erroneous data, and scale this data to accommodate millions of genome sequences.
The Largest-Ever Human Family Tree
Dr. Yan Wong, an evolutionary geneticist at the BDI and one of the principal authors of the study, explains:
“I’m part of this team that assembled this largest-ever human family tree, which included about 3,600 individuals from all over the world. We were able to piece this together into a massive network of links between people – genetic links, using their DNA.”
From eight datasets, the study integrated a total of 3,601 modern human genome sequences and eight high-coverage ancient sequences from 215 populations. The ancient genomes included samples found around the world, ranging in age from a few thousand to over 100,000 years old.
The enormous tree’s branches comprise a mind-blowing 231 million ancestral lineages.
The team focused on DNA fragments that vary from person to person to identify 6,412,717 variants. To explain patterns of genetic variation, the algorithms predicted where common ancestors should be present in evolutionary trees, and the network contained almost 27 million ancestors.
Dr. Wong explains:
“We have essentially built a huge family tree for all of humanity, which models as accurately as possible the history that generated all the genetic variations that we find in humans today. This genealogy allows us to understand how the genetic sequence of each person is linked to all the others, at all points of the genome.”
“The exact way it works is that we sort of try and have a guess at what the genetic ancestors of different sets of people looked like in terms of their DNA sequence at different points in the past.”
“And then, once we’ve had a guess at what those genomic sequences look like, then we can map the sequences that we know about today onto each of those ancient ancestors. And we do that in different places in the genome, this is really key.”
Our oldest roots are in North East Africa
This huge tree also tells us about where people came from – the historical migration roots around the world.
According to the new human family tree, our oldest roots are in northeast Africa. Indeed, the oldest H. sapiens fossils come from northern and eastern Africa, although the original range of our species is still uncertain. The oldest known specimens come from Jebel Irhoud in Morocco, possibly 315,000 years old. It is also possible that there were once several populations spread across the African continent, with very ancient divergences.
Dr. Wilder Wohns, a Postdoctoral Researcher at Broad Institute of MIT and Harvard, explains:
“One of the most interesting results that came out of this work was an insight into where and when human ancestors lived. It’s immediately apparent that human genetic diversity is highest in Africa, as was previously known, and that comes with signals of the ‘Out of Africa’ event.”
“There’s also a great depth of lineages in Oceania which is indicative of complex interactions between the ancestors of people that lived in that part of the world.”
The new tree also modifies our knowledge acquired on the first voyages. While many archaeologists had fixed the oldest entry into the American continent at 18,000 years, the tree seems to indicate that this date would rather be 56,000 years ago. A September 2021 study had also described footprints from White Sands National Park in New Mexico, suggesting a human presence on the American continent 21,000 to 23,000 years ago.
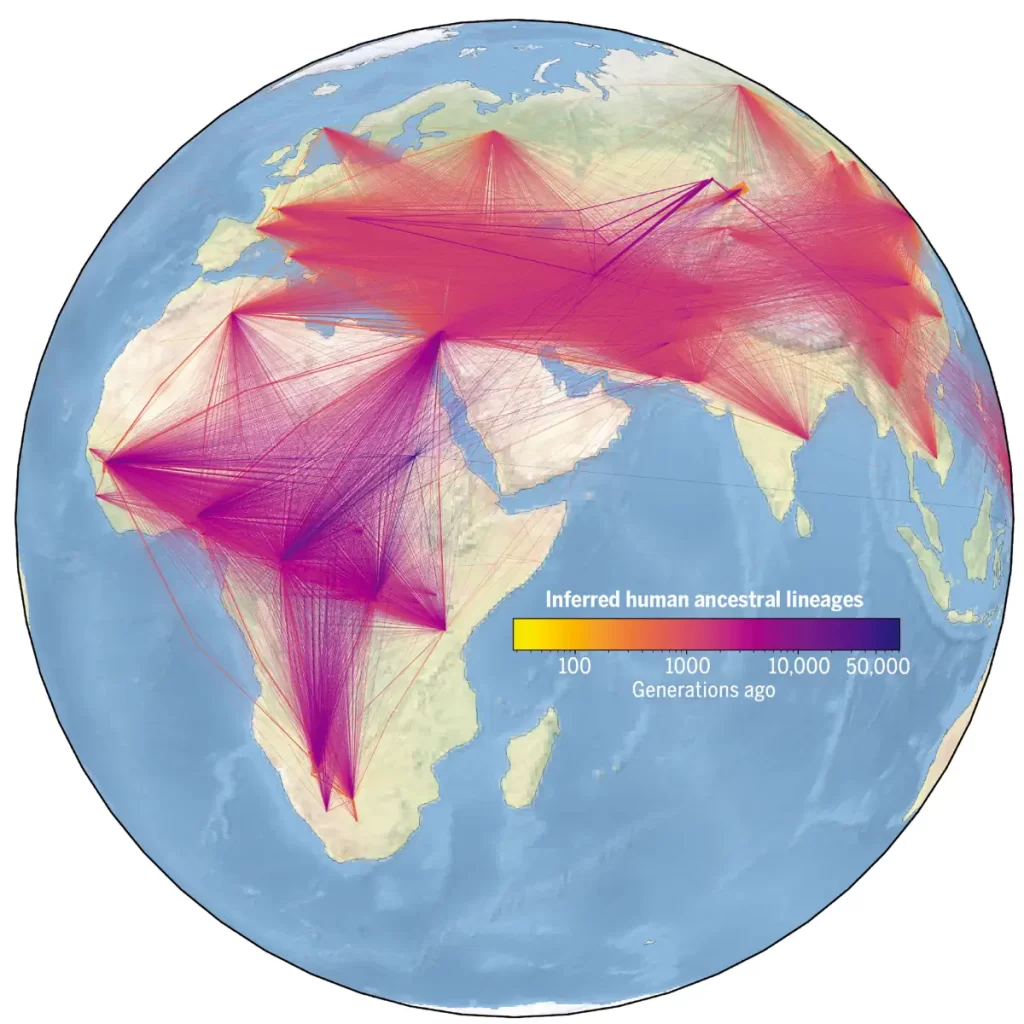
A foundation for future research
The map is already huge, but the research team plans to incorporate more genetic data, likely millions more genomes.
“This study lays the foundation for the next generation of DNA sequencing,” Wong explains. “As the quality of genomic sequences from modern and ancient DNA samples improves, the trees will become even more precise, and we will eventually be able to generate a single, unified map explaining the origin of all the human genetic variations that we observe today”.
Also, the method could apply to other living things, from bacteria to orangutans. It could also have many applications in medical research, for example, to identify genetic predictors of disease risk.
“While humans are the focus of this study, the method is valid for most living things; from orangutans to bacteria,” says Wohns.
Related: Is Evolution “just a theory”?
Sources
- “BDI researchers create largest-ever human family tree” on the Big Data Institute website
- Study: “A unified genealogy of modern and ancient genomes” on Science
- “We Now Have The Largest Ever Human ‘Family Tree’, With 231 Million Ancestral Lineages” on the Science Alert website
- Study: “Evidence of humans in North America during the Last Glacial Maximum” on Science
- Moon Landings: All-Time List [1966-2025] - February 2, 2025
- What Is Max-Q and Why Is It Important During Rocket Launches? - January 16, 2025
- Top 10 Tallest Rockets Ever Launched [2025 Update] - January 16, 2025